Bacterial Engineering
Terra Bioforge Engineers Bacteria For Your Needs
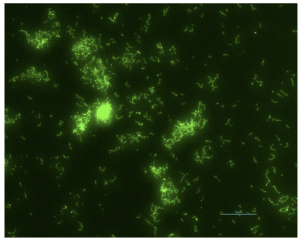
Plant-associated Bacillus stably labeled with green-fluorescent protein for in planta visualization.
At Terra Bioforge, we specialize in engineering bacteria for use in a wide array of beneficial applications including animal health, agriculture, environmental remediation, and human health.
We succeed in engineering strains including Bacillus, Paenibacillus, and Streptomyces, to fine-tune traits that meet your needs.
Select applications include:
- Optimizing plant-growth promotion by modifying nutrient utilization through deletion or up-regulation of target genes
- Stable fluorescent labeling of strains for tracking in vivo or in planta
- Secretion of peptides or small molecules to suppress growth of pathogens or other unwanted members of microbial communities
- Secretion of potent plastic-degrading enzymes for environmental remediation
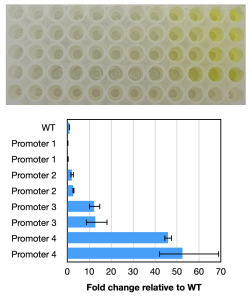
Up to 50-fold improvement in beneficial activity through strain engineering of a plant-associated Bacillus.

High-level expression and secretion of β-agarase in Bacillus.
Contact us at info@terrabioforge.com to discuss your strain engineering needs!
Publications
Publications from customers who used Terra Bioforge technology:
Heterologous expression of the cryptic mdk gene cluster and structural revision of maduralactomycin A
Discovery of the Biosynthetic Machinery for Stravidins, Biotin Antimetabolites DOI
Elucidation of chalkophomycin biosynthesis reveals N-hydroxypyr-role-forming enzymes
Publications from Terra Bioforge team members:
A Survey of Didemnin Depsipeptide Production in Tistrella. Stankey RJ, Johnson D, Duggan BM, Mead DA, La Clair JJ. Marine Drugs. 2023; 21(2):56. https://doi.org/10.3390/md21020056
Correlative metabologenomics of 110 fungi reveals metabolite/gene cluster pairs. Caesar LK, Butun FA, Robey MT, Ayon NJ, Gupta R, Dainko D, Bok JW, Nickles G, Stankey RJ, Johnson D, Mead DA, Cank KB, Earp CE, Raja HA, Oberlies NH, Keller NP, Kelleher NK. Nature Chemical Biology. 2023.
Nanoscaled discovery of a shunt rifamycin from Salinispora arenicola using a three-colour GFP-tagged Staphylococcus aureus macrophage infection assay. Nhan T. Pham, Joana Alves, Fiona A. Sargison, Reiko Cullum, Jan Wildenhain, William Fenical, Mark S. Butler, David A. Mead, Brendan M. Duggan, J. Ross Fitzgerald, James J. La Clair, Manfred Auer. bioRxiv 2022.12.04.519019; doi: https://doi.org/10.1101/2022.12.04.519019
An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Robey MT, Caesar LK, Drott MT, Keller NP, Kelleher NL. Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2020230118. doi: 10.1073/pnas.2020230118. PMID: 33941694; PMCID: PMC8126772.
Discovery of the Biosynthetic Machinery for Stravidins, Biotin Antimetabolites. Montaser R, Kelleher NL. ACS Chem Biol. 2020 May 15;15(5):1134-1140. doi:10.1021/acschembio.9b00890. Epub 2020 Jan 9. PMID: 31887014; PMCID: PMC7230017.
Discovery of Novel Biosynthetic Gene Cluster Diversity from a Soil Metagenomic Library. Santana-Pereira ALR, Sandoval-Powers M, Monsma S, Zhou J, Santos SR, Mead DA, Liles MR. Front Microbiol. 2020 Dec 7;11:585398. doi: 10.3389/fmicb.2020.585398
Chloramphenicol Derivatives with Antibacterial Activity Identified by Functional Metagenomics. Nasrin S, Ganji S, Kakirde KS, Jacob MR, Wang M, Ravu RR, Cobine PA, Khan IA, Wu CC, Mead DA, Li XC, Liles MR. J Nat Prod. 2018 Jun 22;81(6):1321-1332.
A scalable platform to identify fungal secondary metabolites and their gene clusters. Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, Lamprecht M, Thomas PM, Islam MN, Palmer JM, Wu CC, Keller NP, Kelleher NL. Nat Chem Biol. 2017 Aug;13(8):895-901. doi: 10.1038/nchembio.2408. Epub 2017 Jun 12. PMID: 28604695; PMCID: PMC5577364.
Challenges and Opportunities in the Discovery of Secondary Metabolites Using a Functional Metagenomics Approach. Pereira A, Liles MR, In: Functional Metagenomics: Tools and Applications, edited by Charles T, Liles MR, and Sessitch A. pp. 119-138. (2017) Berlin Heidelberg: Springer-Verlag.
Meeting Report for Synthetic Biology for Natural Products 2017: The Interface of (Meta)Genomics, Machine Learning, and Natural Product Discovery. Smanski MJ, Mead D, Gustafsson C, Thomas MG. ACS Synth Biol. 2017 May 19;6(5):737-743.
Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. Goering AW, McClure RA, Doroghazi JR, Albright JC, Haverland NA, Zhang Y, Ju KS, Thomson RJ, Metcalf WW, Kelleher NL. ACS Cent Sci. 2016 Feb 24;2(2):99-108. doi: 10.1021/acscentsci.5b00331. Epub 2016 Jan 20. PMID: 27163034; PMCID: PMC4827660.
Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics. Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Albright JC, Goering AW, Thomas PM, Kelleher NL, Keller NP, Wu CC. 2015 Apr 29;16(1):343. doi: 10.1186/s12864-015-1561-x. PMID: 25925221; PMCID: PMC4413528.
Bacillusin A, an Antibacterial Macrodiolide from Bacillus amyloliquefaciens AP183. Ravu RR, Jacob MR, Chen X, Wang M, Nasrin S, Kloepper JW, Liles MR, Mead DA, Khan IA, Li XC. J Nat Prod. 2015 Apr 24;78(4):924-8.
Draft genome sequence of Bacillus amyloliquefaciens AP183 with antibacterial activity against methicillin-resistant Staphylococcus aureus. Nasrin S, Hossain MJ, and Liles MR (2015) Genome Announcements, 3(2). pii: e00162-15.
A roadmap for natural product discovery based on large-scale genomics and metabolomics. Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. Nat Chem Biol. 2014 Nov;10(11):963-8. doi: 10.1038/nchembio.1659. Epub 2014 Sep 28. PMID: 25262415; PMCID: PMC4201863.
Biological agents for control of disease in aquaculture. Carrias A, Ran C, Terhune J, and Liles MR (2012), pp. 353-393. In: Infectious Diseases in Aquaculture, S. Austin (Ed.). London: Woodhead Publishing.
Recovery of as-yet-uncultured soil Acidobacteria on dilute solid media. George IF, Hartmann M, Liles MR, and Agathos SN. (2011) Applied & Environmental Microbiology, 77(22):8184-8188.
Polyketide synthase pathways identified from a metagenomic library are derived from soil Acidobacteria. Parsley LC, Goode AM, Becklund K, George I, Linneman J, Lopanik NB, Goodman RM, and Liles MR. (2011) FEMS Microbiology Ecology, 78(1):176-187.
Gram-negative shuttle BAC vector for heterologous expression of metagenomic libraries. Kakirde KS, Wild J, Godiska R, Mead D, Wiggins AG, Szybalski W, and Liles MR. (2011) Gene, 475(2):57-62.
Size Does Matter: Application-driven Approaches for Soil Metagenomics. Kakirde KS, Parsley LC, and Liles MR. (2010) Soil Biology and Biochemistry, 42(11):1911-1923.
Isolation and cloning of high molecular weight metagenomic DNA from soil microorganisms. Liles M R, Williamson LL, Rodbumrer J, Parsley L, Torsvik V, Goodman RM, and Handelsman J. (2009) Cold Spring Harbor Protocols, doi:10.1101/pdb.prot5271.
Recovery, purification, and cloning of high molecular weight genomic DNA from soil microorganisms. Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, and Handelsman J. (2008) Applied and Environmental Microbiology, 74:3302-3305.
Isolation of high molecular weight genomic DNA from soil bacteria for genomic library construction. Liles MR, Williamson LL, Goodman RM, and Handelsman J. (2004) In: Molecular Methods in Environmental Microbiology, de Bruijn FJ, Head IM, Akkermans AD, van Elsas JD. (Eds.), pp. 839-852.
Cloning the metagenome: Culture-independent access to the diversity and functions of the uncultivated microbial world. Handelsman J, Liles MR, Mann D, Riesenfeld C, and Goodman RM. (2002) Methods in Microbiology, 33:241-255.
Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. (2002). Applied and Environmental Microbiology, 68:4301-4306.
Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osbourne MS, Clardy J, Handelsman J, and Goodman RM (2000) Applied and Environmental Microbiology, 66 (6): 2541-2547.
Interested in learning more about science and technology at Terra Bioforge? Please contact us.