Core Technologies
We have built a platform to discover and manufacture valuable natural products from fungi and other microbes. Our technology makes this process 10-100 times faster than traditional methods, which means we can bring new products to market more quickly and at a lower cost.
Please contact us at info@terrabioforge.com for technical information or to request a quote for Terra Bioforge’s research and lab services.
MycoDrive™
A Breakthrough Platform to Accelerate High-Yield Fungal Production
MycoDrive™ is a next-generation genome engineering platform that turns filamentous fungi into ultra-efficient production hosts—delivering dramatically higher yields in a fraction of the time.
For markets where production output determines whether a product can scale or succeed, MycoDrive unlocks the high titers needed to accelerate development, reduce costs, and bring valuable fungal innovations to market faster.
Applications & Benefits:
- Boost product yields for natural products, BGCs*, and enzymes — enabling products that were previously too low-producing to be viable
- Lower manufacturing costs (COGS) by creating smarter, more efficient production strains
- Engineer in the best host for the job — whether improving the native organism or moving production into a faster, easier-to-scale microbe
- Accelerate development timelines with more high-performing strains available for rapid testing and scale-up

-
*A Biosynthetic Gene Cluster (BGC) is a compact set of genes inside microbes that functions like a ready-made manufacturing line. Each cluster produces a specific high-value product — such as a therapeutic, bioactive compound, enzyme, food ingredient, or material — and can be switched on, optimized, or repurposed for commercial use.
-
Why this matters: MycoDrive technology allows companies to rapidly turn microbes into scalable production systems, shortening development timelines and enabling new product pipelines in health, food, and sustainability markets.
-
- *AMA1 is a DNA element that allows fungi to rapidly and flexibly express multiple copies of new genes — enabling faster strain development, higher product yields, and lower commercialization risk.
-
Why this matters: AMA1 accelerates development timelines, increases production yields, and reduces the cost and risk of bringing new fungal-based products to market—directly improving return on R&D and commercialization success rates. MycoDrive technology leapfrogs AMA1 plasmids by providing long-term genome integration stability and higher copy numbers of BGCs.
-
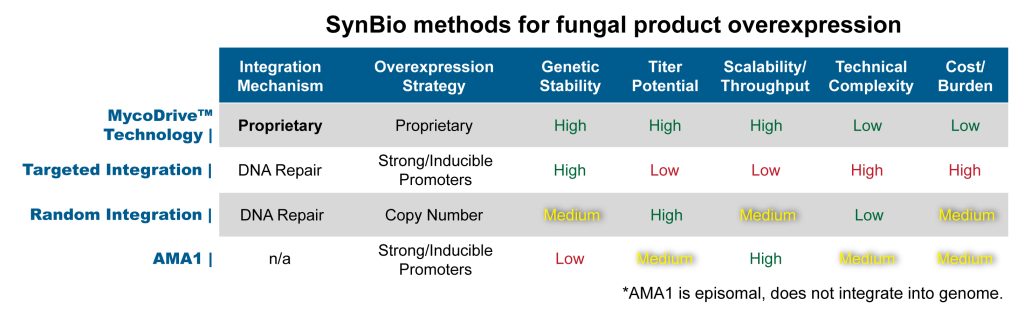
CASE STUDY – Heterologous Overproduction of Bikaverin
Bikaverin is a multi-gene purple pigment used to visualize expression strength. It’s pathway from Fusarium oxysporum was heterologously expressed in Aspergillus nidulans using MycoDrive technology to attain expression titers ~30x higher over incumbent genome engineering methods.
Ongoing work is expanding this transformative tool into new fungal hosts and further boosting native or heterologous production titers which will enable a shift in expectations for what synthetic biology can do for industrial mycology.

MycoDrive technology quickly attains highest titer
- Supplements BGC Refactoring
- BGC refactoring is often necessary to convert a poorly expressed gene cluster into a high-performance manufacturing module suitable for scalable bioproduction.
- Straightforward cloning – no HOM arms or dealing with AMA1 repeats
- HOM arms or Homology arms are used for homologous recombination during CRISPR editing — they show the cell exactly where to place new DNA, enabling accurate and efficient genome engineering.
- Heterologous titer in A. nidulans can beat native producer F. oxysporum
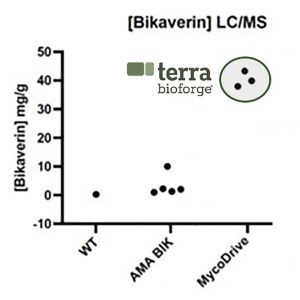
BGC Copy Number Positively Correlates with Heterologous Bikaverin
Over half of transformants generated with MycoDrive were high bikaverin producers. To better understand the relationship between BGC copy number and heterologous natural product production, digital PCR (dPCR) was used to quantify BGC copy number/genome. Integration frequency correlates with increasing bikaverin production up to ~5-10 copies/genome. More than that and, not surprisingly, there appears to be another production bottleneck which could be overcome with targeted genetic manipulations, process engineering, or even mutagenesis.
Random integration methods can oftentimes require screening 1,000s of transformants to find stable strains with intact payloads and high performance. MycoDrive engineering produces high performing clones early in the R&D process.
What can MycoDrive engineering do for you?

Let’s discuss how MycoDrive technology can advance your fungal product R&D and production!
Benjamin Knox, PhD – Director of Fungal Technology bknox@terrabioforge.com https://vn7qtk8lp41.c.updraftclone.com/
BIGDNA®
Tools for Manipulating BGCs
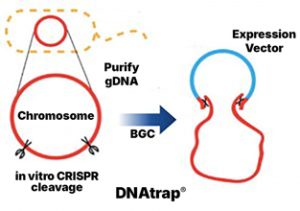
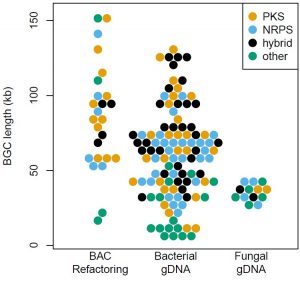 Natural products are made from recipes encoded with BGCs; historically, it has been very difficult to work with these large pieces of DNA.
Natural products are made from recipes encoded with BGCs; historically, it has been very difficult to work with these large pieces of DNA.
Our BIGDNA® tools, including DNAtrap®, allow us to capture and modify these entire recipes from any microbe, which is a critical step in developing new products.
- We can clone very large gene clusters (up to 150 kb) that are usually ‘impossible’ to move because they’re repetitive or GC-rich
- Effectively captures BGCs from complex genomic DNA (gDNA) or metagenomic library (DNA from uncultured environmental microbes, where many of the best natural products hide) substrates
- De-Risked: DNAtrap® has been used to clone >130 intact BGCs to date
Foundational Technologies for Complex Molecule Manufacturing
BAC (Bacterial Artificial Chromosome)
A large-capacity, stable DNA carrier that enables companies to move and express massive, high-value genetic programs—such as entire natural product pathways—inside production strains.
PKS (Polyketide Synthase)
A modular biosynthesis engine used by microbes to create valuable small-molecule products, including many blockbuster drugs, with tunable architectures that support rapid innovation.
NRPS (Nonribosomal Peptide Synthetase)
A natural molecular foundry that assembles complex therapeutic and industrial peptides with precision—unlocking access to differentiated products that can’t be made by standard biology.
Hybrid PKS–NRPS Systems
Integrated biosynthetic platforms that combine the strengths of both PKS and NRPS machinery, enabling discovery and scalable manufacturing of uniquely complex, high-margin molecules.
Engineering Expression of BGCs
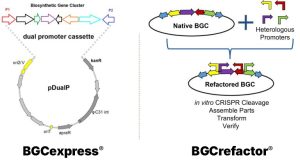 In both native and heterologous microbial hosts natural product production is an innately regulated process. These pathways are often naturally ‘switched off’ or very low, which makes their products hard to detect, measure, or optimize.
In both native and heterologous microbial hosts natural product production is an innately regulated process. These pathways are often naturally ‘switched off’ or very low, which makes their products hard to detect, measure, or optimize.
Our team has pioneered novel methodologies to predictably activate BGCs for both natural product screening and titer optimization workflows.
- BGCexpress® allows you to “switch on” natural product pathways only when needed, boosting yields while maintaining healthy growth.
- The result: 2–3× higher titers that make it easier to identify promising molecules and advance the best candidates quickly.
- BGCrefactor® streamlines the redesign of entire biosynthetic gene clusters—quickly removing genetic roadblocks and enabling higher-performing production strains.
- The result: A multiplexed, marker-free workflow simplifies pathway optimization end-to-end, accelerating the creation of improved natural product titers and more scalable biomanufacturing.
Together, these tools unlock previously silent or low-yield pathways and convert them into highly productive, tunable manufacturing systems—significantly accelerating optimization and time-to-value.
Metabologenomics for Natural Product BGC Identification
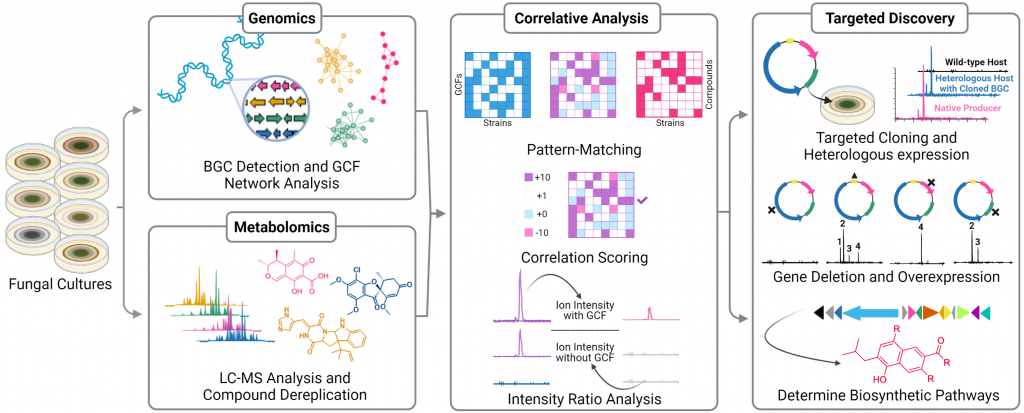 Metabologenomics is a data-driven method that links what an organism can produce (from its genome) with what it actually produces (its metabolites). By combining genomic sequencing and metabolite profiling, it rapidly reveals which metabolic pathways are active and which ones could be turned into new, valuable products — like breakthrough drugs, enzymes, or specialty chemicals.
Metabologenomics is a data-driven method that links what an organism can produce (from its genome) with what it actually produces (its metabolites). By combining genomic sequencing and metabolite profiling, it rapidly reveals which metabolic pathways are active and which ones could be turned into new, valuable products — like breakthrough drugs, enzymes, or specialty chemicals.
In short, Metabologenomics turns biological “blueprints” into commercial discovery opportunities faster and more efficiently.
One of the biggest barriers in natural product discovery is knowing which genes create which valuable molecules. Our patented Metabologenomics platform removes that guesswork. By combining advanced bioinformatics with AI-driven metabolite analysis, we can map every molecule a microbe makes back to its exact genetic “recipe”. This gives us a fast, reliable way to uncover previously hidden natural products — and then engineer them into scalable, commercially viable assets.
- Prospect Fungi is a proprietary analysis tool for in silico prediction and annotation of fungal BGCs
- An AI platform that grows and learns from known BGC: Natural Product relationships
- Haystack-MS: Finding new drugs via targeted cloning
- Solves needle-in-the-haystack problem of finding target compounds in complex metabolite mixtures
- Rapidly identifies and produces natural products by heterologous expression
We’ve essentially automated the critical step of linking biosynthetic gene clusters to commercially valuable molecules, transforming trial-and-error discovery into a predictable, accelerated pipeline that reduces development risk.
Hosts for Heterologous Expression
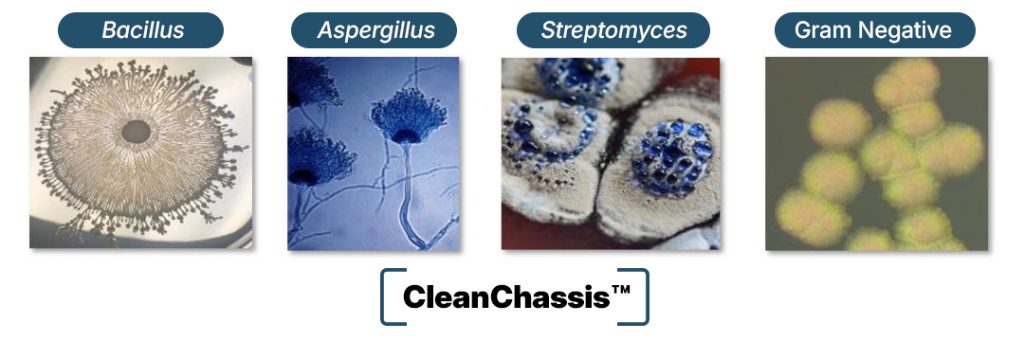 When it comes to optimal heterologous expression of natural products, host selection matters. Precursor metabolites, cofactors, among other host-specific variables can greatly influence the successful production of specific molecules. Our access to a suite of optimal heterologous hosts provides the greatest opportunity for natural product expression.
When it comes to optimal heterologous expression of natural products, host selection matters. Precursor metabolites, cofactors, among other host-specific variables can greatly influence the successful production of specific molecules. Our access to a suite of optimal heterologous hosts provides the greatest opportunity for natural product expression.
- CleanChassis™ strains utilizing the main industrial workhorses used in pharma, agriculture, and enzymes, have been engineered to reduce competing metabolite production:
- Streptomyces, Bacillus, Aspergillus, and Gram Negative bacteria
- For every BGC of interest, our growing portfolio of CleanChassis™ strains represents an opportunity to accelerate optimal expression
For additional information on Terra Bioforge technical capabilities, check out our conference posters on New Tools for Targeted Cloning and Over Expression of Biosynthetic Gene Clusters and Expanding Functional Access to Fungal Natural Products with Metabologenomics and Heterologous Expression.
Publications
Publications from customers who used Terra Bioforge technology:
Heterologous expression of the cryptic mdk gene cluster and structural revision of maduralactomycin A
Discovery of the Biosynthetic Machinery for Stravidins, Biotin Antimetabolites DOI
Elucidation of chalkophomycin biosynthesis reveals N-hydroxypyr-role-forming enzymes
Publications from Terra Bioforge team members:
A Survey of Didemnin Depsipeptide Production in Tistrella. Stankey RJ, Johnson D, Duggan BM, Mead DA, La Clair JJ. Marine Drugs. 2023; 21(2):56. https://doi.org/10.3390/md21020056
Correlative metabologenomics of 110 fungi reveals metabolite/gene cluster pairs. Caesar LK, Butun FA, Robey MT, Ayon NJ, Gupta R, Dainko D, Bok JW, Nickles G, Stankey RJ, Johnson D, Mead DA, Cank KB, Earp CE, Raja HA, Oberlies NH, Keller NP, Kelleher NK. Nature Chemical Biology. 2023.
Nanoscaled discovery of a shunt rifamycin from Salinispora arenicola using a three-colour GFP-tagged Staphylococcus aureus macrophage infection assay. Nhan T. Pham, Joana Alves, Fiona A. Sargison, Reiko Cullum, Jan Wildenhain, William Fenical, Mark S. Butler, David A. Mead, Brendan M. Duggan, J. Ross Fitzgerald, James J. La Clair, Manfred Auer. bioRxiv 2022.12.04.519019; doi: https://doi.org/10.1101/2022.12.04.519019
An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Robey MT, Caesar LK, Drott MT, Keller NP, Kelleher NL. Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2020230118. doi: 10.1073/pnas.2020230118. PMID: 33941694; PMCID: PMC8126772.
Discovery of the Biosynthetic Machinery for Stravidins, Biotin Antimetabolites. Montaser R, Kelleher NL. ACS Chem Biol. 2020 May 15;15(5):1134-1140. doi:10.1021/acschembio.9b00890. Epub 2020 Jan 9. PMID: 31887014; PMCID: PMC7230017.
Discovery of Novel Biosynthetic Gene Cluster Diversity from a Soil Metagenomic Library. Santana-Pereira ALR, Sandoval-Powers M, Monsma S, Zhou J, Santos SR, Mead DA, Liles MR. Front Microbiol. 2020 Dec 7;11:585398. doi: 10.3389/fmicb.2020.585398
Chloramphenicol Derivatives with Antibacterial Activity Identified by Functional Metagenomics. Nasrin S, Ganji S, Kakirde KS, Jacob MR, Wang M, Ravu RR, Cobine PA, Khan IA, Wu CC, Mead DA, Li XC, Liles MR. J Nat Prod. 2018 Jun 22;81(6):1321-1332.
A scalable platform to identify fungal secondary metabolites and their gene clusters. Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, Lamprecht M, Thomas PM, Islam MN, Palmer JM, Wu CC, Keller NP, Kelleher NL. Nat Chem Biol. 2017 Aug;13(8):895-901. doi: 10.1038/nchembio.2408. Epub 2017 Jun 12. PMID: 28604695; PMCID: PMC5577364.
Challenges and Opportunities in the Discovery of Secondary Metabolites Using a Functional Metagenomics Approach. Pereira A, Liles MR, In: Functional Metagenomics: Tools and Applications, edited by Charles T, Liles MR, and Sessitch A. pp. 119-138. (2017) Berlin Heidelberg: Springer-Verlag.
Meeting Report for Synthetic Biology for Natural Products 2017: The Interface of (Meta)Genomics, Machine Learning, and Natural Product Discovery. Smanski MJ, Mead D, Gustafsson C, Thomas MG. ACS Synth Biol. 2017 May 19;6(5):737-743.
Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer. Goering AW, McClure RA, Doroghazi JR, Albright JC, Haverland NA, Zhang Y, Ju KS, Thomson RJ, Metcalf WW, Kelleher NL. ACS Cent Sci. 2016 Feb 24;2(2):99-108. doi: 10.1021/acscentsci.5b00331. Epub 2016 Jan 20. PMID: 27163034; PMCID: PMC4827660.
Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics. Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Albright JC, Goering AW, Thomas PM, Kelleher NL, Keller NP, Wu CC. 2015 Apr 29;16(1):343. doi: 10.1186/s12864-015-1561-x. PMID: 25925221; PMCID: PMC4413528.
Bacillusin A, an Antibacterial Macrodiolide from Bacillus amyloliquefaciens AP183. Ravu RR, Jacob MR, Chen X, Wang M, Nasrin S, Kloepper JW, Liles MR, Mead DA, Khan IA, Li XC. J Nat Prod. 2015 Apr 24;78(4):924-8.
Draft genome sequence of Bacillus amyloliquefaciens AP183 with antibacterial activity against methicillin-resistant Staphylococcus aureus. Nasrin S, Hossain MJ, and Liles MR (2015) Genome Announcements, 3(2). pii: e00162-15.
A roadmap for natural product discovery based on large-scale genomics and metabolomics. Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. Nat Chem Biol. 2014 Nov;10(11):963-8. doi: 10.1038/nchembio.1659. Epub 2014 Sep 28. PMID: 25262415; PMCID: PMC4201863.
Biological agents for control of disease in aquaculture. Carrias A, Ran C, Terhune J, and Liles MR (2012), pp. 353-393. In: Infectious Diseases in Aquaculture, S. Austin (Ed.). London: Woodhead Publishing.
Recovery of as-yet-uncultured soil Acidobacteria on dilute solid media. George IF, Hartmann M, Liles MR, and Agathos SN. (2011) Applied & Environmental Microbiology, 77(22):8184-8188.
Polyketide synthase pathways identified from a metagenomic library are derived from soil Acidobacteria. Parsley LC, Goode AM, Becklund K, George I, Linneman J, Lopanik NB, Goodman RM, and Liles MR. (2011) FEMS Microbiology Ecology, 78(1):176-187.
Gram-negative shuttle BAC vector for heterologous expression of metagenomic libraries. Kakirde KS, Wild J, Godiska R, Mead D, Wiggins AG, Szybalski W, and Liles MR. (2011) Gene, 475(2):57-62.
Size Does Matter: Application-driven Approaches for Soil Metagenomics. Kakirde KS, Parsley LC, and Liles MR. (2010) Soil Biology and Biochemistry, 42(11):1911-1923.
Isolation and cloning of high molecular weight metagenomic DNA from soil microorganisms. Liles M R, Williamson LL, Rodbumrer J, Parsley L, Torsvik V, Goodman RM, and Handelsman J. (2009) Cold Spring Harbor Protocols, doi:10.1101/pdb.prot5271.
Recovery, purification, and cloning of high molecular weight genomic DNA from soil microorganisms. Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, and Handelsman J. (2008) Applied and Environmental Microbiology, 74:3302-3305.
Isolation of high molecular weight genomic DNA from soil bacteria for genomic library construction. Liles MR, Williamson LL, Goodman RM, and Handelsman J. (2004) In: Molecular Methods in Environmental Microbiology, de Bruijn FJ, Head IM, Akkermans AD, van Elsas JD. (Eds.), pp. 839-852.
Cloning the metagenome: Culture-independent access to the diversity and functions of the uncultivated microbial world. Handelsman J, Liles MR, Mann D, Riesenfeld C, and Goodman RM. (2002) Methods in Microbiology, 33:241-255.
Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. (2002). Applied and Environmental Microbiology, 68:4301-4306.
Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osbourne MS, Clardy J, Handelsman J, and Goodman RM (2000) Applied and Environmental Microbiology, 66 (6): 2541-2547.
Interested in learning more about science and technology at Terra Bioforge? Please contact us.